What is Clustering?
Unsupervised clustering algorithms are commonly used in areas such as genomics, machine learning, pattern recognition, and data compression to divide a set of unlabeled observations into separate groups with similar traits.
The most widely used partitional clustering algorithm is K-Means. This algorithm is commonly used not only on its own, but also as a component of ensemble clustering.
Mini Batch K-Means Clustering Algorithm
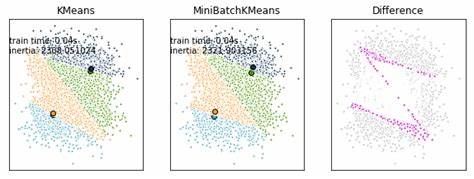
K-Means is one of the most used clustering algorithms, mainly because of its good time perforamance. With the increasing size of the datasets being analyzed, this algorithm is losing its attractive because its constraint of needing the whole dataset in main memory. For this reason several methods have been proposed to reduce the temporal and spatial cost of the algorithm.
For example, in it is proposed a modification in how is computed the assignment of examples to cluster prototypes using the triangular inequality. This method effectively reduces the number of distance computations each iteration, but maintaining the need of having all the dataset in memory. In several strategies are used to reduce the amount of data needed to update the cluster centroids each iteration by selecting random samples of the dataset, by summarizing examples using sufficient statistics and by discarding examples that have a small impact on the clusters.
A different approach is the mini batch K-means algorithm. Its main idea is to use small random batches of examples of a fixed size so they can be stored in memory. Each iteration a new random sample from the dataset is obtained and used to update the clusters and this is repeated until convergence. Each mini batch updates the clusters using a convex combination of the values of the prototypes and the examples, applying a learning rate that decreases with the number of iterations.
This learning rate is the inverse of number of examples assigned to a cluster during the process. As the number of iterations increases, the effect of new examples is reduced, so convergence can be detected when no changes in the clusters occur in several consecutive iterations.
Mini Batch K-Means has been proposed as an alternative to the K-Means algorithm for clustering massive datasets. The advantage of this algorithm is to reduce the computational cost by not using all the dataset each iteration but a subsample of a fixed size. This strategy reduces the number of distance computations per iteration at the cost of lower cluster quality.
K-Means algorithm can be broken down into five steps
- Place all instances into subsets, where the number of subsets is equal to K.
- Find the mean centroid of the created cluster partitions.
- Based on these centroids, assign each point to a specific cluster.
- Calculate the distances from every point to the centroids, and assign points to the clusters where the distance from centroid is the minimum.
- After the points have been assigned to the clusters, find the new centroid of the clusters.
Choose the Right Value for ‘K’
One technique for selecting the right K value is called the “the elbow technique”. This technique consists of running a K-Means algorithm for a range of different K values and using an accuracy metric (sum of squared error), to determine which values of K give the best results. The is determined by calculating the mean distance between the centroid of a cluster and the data points in that cluster.
Elbow technique comes from the fact that when you plot the SSE with regard to the different values of K, the resulting line plot will often have an elbow shape, where accuracy decreases rapidly for the first few values of K, but then levels off. In such conditions, the value of K located at the elbow is the best value for K.
Another technique to get the optimal value of K is to use the autooptimizer package. Using combined techniques of Exhaustive Search and Autotuning , the library obtains the best possible K value based on the dataset.
Overview of Mini Batch K-Means Algorithm
While K-Means is widely used, two problems with its implementation are it requires all the data to be stored in memory and it may be computationally expensive when the sample size or the number of clusters are large. Stochastic gradient descent methods compute a gradient descent step one observation at a time, and are often used to improve the speed and memory usage. The primary disadvantage is they find typically lower quality solutions due to stochastic noise; a standard solution is to use batches of data at each gradient descent, rather than individual data points (mini batch gradient descent).
This strategy was proposed in the context of K-Means for improving its computational performance, and is known as the Mini Batch K-Means algorithm. The use of mini-batches has been shown to have lower stochastic noise without the expensive computational time in the K-Means algorithm with large datasets. The use of the mini batches also allows for the ability to not store the entire dataset in memory. Therefore, The Mini Batch K-Means algorithm is an ideal approach to allow for fast, scalable, memory-efficient clustering data.
Properties of K-Means
- No need to store the whole dataset in the memory.
- At each iteration, distance between the mini-batch and the k centroids needs to be calculated.
- At each iteration, user needs to store k centroids and a subset of data in the memory.
Recommended for you:




MOST COMMENTED
Tutorial
Important Methods in Matplotlib
Machine Learning
Bias and Variance Tradeoff Machine Learning
Tutorial
Multiclass and Multilabel Classification
Machine Learning
Reinforcement Learning in Machine Learning
Deep Learning
Alexnet Architecture Code
Machine Learning
Machine Learning Models Explained